make?make is a command-line program of the same pedigree & importance as classic UNIX programs like grep and ssh.
A powerful tool that has stood the test of time, make is readily available in Linux and MacOS as a shell program. Originally used as a build automation tool for making files, make can be used with any workflow that involves running shell programs.
This post shows how to use classic tool in a modern data science project:
Makefile in a data science project,Makefile,Makefile to ingest and clean data.make for Data Science?make is a great tool for data science projects - it provides:
Documenting your project is a basic quality of a good data project.
A Makefile serves as executable documentation of what your project does. Like any text file it’s easy to track changes in source control.
Creating your pipeline as a sequence of make targets also naturally drives your pipelines to be modular - encouraging good functional decomposition of shell or Python scripts.
A Makefile tightly integrates with the shell. We can easily configure variables at runtime via either shell environment variables or via command line arguments.
The Makefile below has two variables - NAME and COUNTRY:
.PHONY: all
all:
echo "$(NAME) is from $(COUNTRY)"
We can set variables using two different methods:
EXPORT name=adam - setting our variable NAME by to a shell environment variable,COUNTRY=NZ - our COUNTRY variable by an argument passed to the make command.$ export NAME=adam; make COUNTRY=NZ
echo "$(NAME) is from $(COUNTRY)"
adam is from nz
We can also assign the output values of shell commands using:
VAR = $(shell echo value)
We have already seen the functionality of intelligent pipeline re-execution - it’s a powerful way to not re-run code that doesn’t need to run.
make uses timestamps on files to track what to re-run (or not re-run) - it won’t re-run code that has already been run and will re-run if dependencies of the target change.
This can save you lots of time - not rerunning that expensive data ingestion and cleaning step when you are working on model selection.
Makefile?A Makefile has three components:
PHONY target,TAB separated steps.target: dependencies
echo "any shell command"
Much of the power of a Makefile comes from being able to introduce dependencies between targets.
The simple Makefile below shows a two step pipeline with one dependency - our run target depends on the setup target:
# define our targets as PHONY as they do not create files
.PHONY: setup run
setup:
pip install -r requirements.txt
run: setup
python main.py
This ability to add dependencies between targets allows a Makefile to represent complex workflows, including data science pipelines.
MakefileThe Makefile below creates an empty data.html file:
data.html:
echo "making data.html"
touch data.html
Running $ make without a target will run the first target - in our case the only target, data.html.
make prints out the commands as it runs:
$ make
making data.html
touch data.html
If we run this again, we see that make runs differently - it doesn’t make data.html again:
$ make
make: `data.html' is up to date.
If we do reset our pipeline (by deleting data.html), running make will run our pipeline again:
$ rm data.html; make
making data.html
touch data.htmlThis is make intelligently re-executing of pipeline.
Under the hood, make makes use of the timestamps on files to understand what to run (or not run).
make Data Pipeline ExampleWe will build a data pipeline - using Python scripts as mock for real data tasks - with data flowing from left to right.
Our ingestion step creates raw data, and our cleaning step creates clean data:
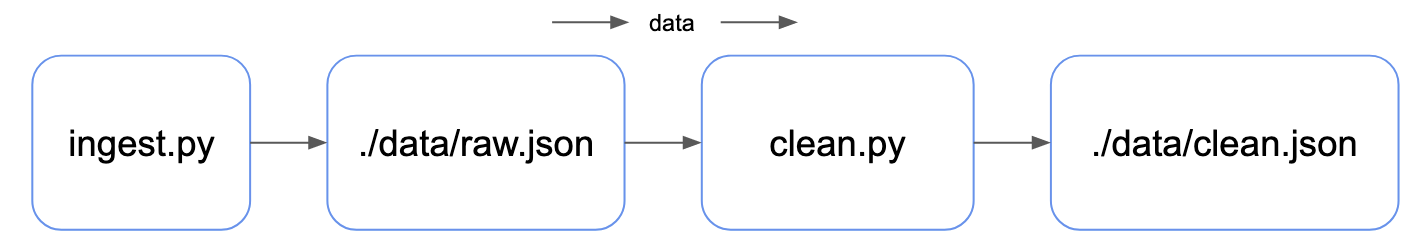
We can look at the same pipeline in terms of the dependency between the data artifacts & source code of our pipeline - with dependency flowing from right to left:
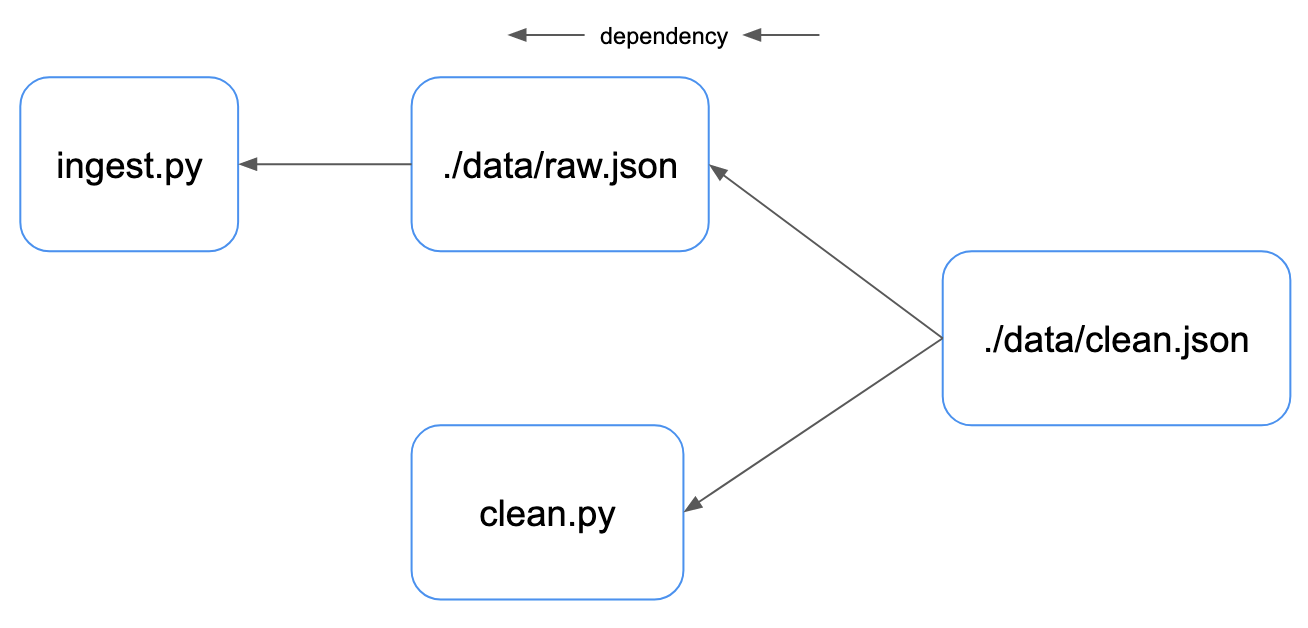
Our clean data depends on both the code used to generate it and the raw data. Our raw data depends only on the ingestion Python script.
Lets look at the two components in our pipeline - an ingestion step and a cleaning step, both of which are Python scripts.
ingest.py writes some data to a JSON file:
#!/usr/bin/env python3
from datetime import datetime
import json
from pathlib import Path
fi = Path.cwd() / "data" / "raw.json"
fi.parent.mkdir(exist_ok=True)
fi.write_text(json.dumps({"data": "raw", "ingest-time": datetime.utcnow().isoformat()}))We can run this Python script and use cat to take a look at it’s JSON output:
$ ./ingest.py; cat data/raw.json
{"data": "raw", "ingest-time": "2021-12-19T13:57:53.407280"}clean.py takes the raw data generated and updates the data field to clean:
#!/usr/bin/env python3
from datetime import datetime
import json
from pathlib import Path
data = json.loads((Path.cwd() / "data" / "raw.json").read_text())
data["data"] = "clean"
data["clean-time"] = datetime.utcnow().isoformat()
fi = Path.cwd() / "data" / "clean.json"
fi.write_text(json.dumps(data))We can use cat again to look at the result of our cleaning step:
$ ./clean.py; cat data/clean.json
{"data": "clean", "ingest-time": "2021-12-19T13:57:53.407280", "clean-time": "2021-12-19T13:59:47.640153"
Let’s start out with a Makefile that runs our two stage data pipeline.
We are already taking advantage of the ability to create dependencies between our pipeline stages, making our clean target depend on our raw target.
We have also included a top level meta target all which depends on our clean step:
.PHONY: all raw clean
all: clean
raw:
mkdir -p data
./ingest.py
clean: raw
./clean.py
We can use this Makefile from a terminal using by running make, which will run our meta target all:
$ make
mkdir -p data
./ingest.py
ingesting {'data': 'raw', 'ingest-time': '2021-12-19T14:14:54.765570'}
./clean.py
cleaning {'data': 'clean', 'ingest-time': '2021-12-19T14:14:54.765570', 'clean-time': '2021-12-19T14:14:54.922659'}
If we go and run only the clean step of our pipeline, we run both the ingest and cleaning step again. This is because our cleaning step depends on the output of data ingestion:
$ make clean
mkdir -p data
./ingest.py
ingesting {'data': 'raw', 'ingest-time': '2021-12-19T14:15:21.510687'}
./clean.py
cleaning {'data': 'clean', 'ingest-time': '2021-12-19T14:15:21.510687', 'clean-time': '2021-12-19T14:15:21.667561'}
What if we only want to re-run our cleaning step? Our next Makefile iteration will avoid this unnecessary re-execution.
Now let’s improve our Makefile, by making changing our targets to be actual files - the files generated by that target.
.PHONY: all
all: clean
./data/raw.json:
mkdir -p data
./ingest.py
./data/clean.json: ./data/raw.json
./clean.py
Removing any output from previous runs with rm -rf ./data, we can run full our pipeline with make:
$ rm -rf ./data; make
mkdir -p data
./ingest.py
ingesting {'data': 'raw', 'ingest-time': '2021-12-27T13:56:30.045009'}
./clean.py
cleaning {'data': 'clean', 'ingest-time': '2021-12-27T13:56:30.045009', 'clean-time': '2021-12-27T13:56:30.193770'}
Now if we run make a second time, nothing happens:
$ make
make: Nothing to be done for `all'.
If we do want to only re-run our cleaning step, we can remove the previous output and run our pipeline again - with make knowing that it only needs to run the cleaning step again with existing raw data:
$ rm ./data/clean.json; make
./clean.py
cleaning {'data': 'clean', 'ingest-time': '2021-12-27T13:56:30.045009', 'clean-time': '2021-12-27T14:02:30.685974'}
The final improvement we will make to our pipeline is to track dependencies on source code.
Let’s update our clean.py script to also track clean-date:
#!/usr/bin/env python3
from datetime import datetime
import json
from pathlib import Path
data = json.loads((Path.cwd() / "data" / "raw.json").read_text())
data["data"] = "clean"
data["clean-time"] = datetime.utcnow().isoformat()
data["clean-date"] = datetime.utcnow().strftime("%Y-%m-%d")
fi = Path.cwd() / "data" / "clean.json"
fi.write_text(json.dumps(data))And now our final pipeline:
all: ./data/clean.json
./data/raw.json: ./data/raw.json ./ingest.py
mkdir -p data
./ingest.py
./data/clean.json: ./data/raw.json ./clean.py
./clean.py
Our final step, after updating only our clean.py script, make will run our cleaning step again:
$ make
./clean.py
ingesting {'data': 'clean', 'ingest-time': '2021-12-27T13:56:30.045009', 'clean-time': '2021-12-27T14:10:06.799127', 'clean-date': '2021-12-27'}
That’s it! We hope you have enjoyed learning a bit about make & Makefile, and are enthusiastic to experiment with it in your data work.
There is more depth and complexity to make and the Makefile - what you have seen so far is hopefully enough to encourage you to experiment and learn more while using a Makefile in your own project.
Key takeaways are:
make is a powerful, commonly available tool that can run arbitrary shell workflows,Makefile forms a natural central point of execution for a project, with a simple CLI that integrates well with the shell,make can intelligently re-execute your data pipeline - keeping track of the dependencies between code and data.